
De Novo Assembly Geneious Microbial de novo Assembly for Illumina Data Introductory Tutorial. Written and maintained by Simon Gladman - Melbourne Bioinformatics (formerly VLSCI)
What Is The Most Memory-Efficient De Novo Assembler?
de Novo assembly Galaxy. ABySS De novo assembly of Illumina reads using ABySS and alignment using BWA. This workshop is designed by Shaun Jackman @sjackman. Purpose. We will use ABySS to, The goal of this tutorial is to gain familiarity with a commonly used procedure for de novo whole genome assembly of For this tutorial the assembler.
Genome Assembly and Editing. The Genomics Suite allows you to quickly and easily perform de novo genome assemblies from any sequencing platform. StringTie is a fast and highly efficient It uses a novel network flow algorithm as well as an optional de novo assembly step to assemble and quantitate full
Whole Genome Assembly and Alignment Assembly Tutorial . Ingredients for a good assembly Current challenges in de novo plant genome sequencing and assembly This tutorial illustrates how to import FASTQ files into a BioNumerics database, how to obtain a quick overview of the basic statistics on read quality and read
Tutorial De Novo Assembly and BLAST 3 After a short while, the reads will be imported. Assembly The reads we are using in this tutorial are on average around 235 bp long. De novo genome assembly Dr Torsten Seemann De novo assembly The process of reconstructing Online tutorial
I've been thinking the SOAPdenovo is the most memory-efficient de novo assembler, but my server can't assemble using SOAPdenovo. I guess RAM capacity is not sufficient. Tutorial De Novo Assembly and BLAST 3 After a short while, the reads will be imported. Assembly The reads we are using in this tutorial are on average around 235 bp long.
transabyss - de novo assembly of RNA-seq data using ABySS 1 Please use To actually complete this tutorial, go to the RNA-seq tutorial wiki. bcbio-nextgen The goal of this tutorial is to gain familiarity with a commonly used procedure for de novo whole genome assembly of For this tutorial the assembler
Assembly using Velvet. Keywords: de novo assembly, Velvet, Galaxy, Microbial Genomics Virtual Lab. Background. Velvet is one of a number of de novo assemblers that TTutorialutorial Tutorial: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45
NGS Pre-Processing and De Novo Assembly Tutorial. In this tutorial you will learn how to process and de novo assemble next-generation sequencing data. Using the Velvet de novo assembler for short-read sequencing technologies. The ABI de novo tools Wang J. De novo assembly of human genomes with massively
4.2.3. Details¶ Snakemake de-novo assembly pipeline dedicates to small genome like bacteria. It bases on SPAdes. The assembler corrects reads then assemble them 10.3 De novo assembly 13.2 Manual primer design under the Help menu in Geneious provides an inbuilt tutorial with a
This tutorial is targeted at scientists with a background in Most other strategies for de novo assembly of short sequence reads can be broadly divided Trinity RNA-Seq de novo transcriptome assembly. Contribute to trinityrnaseq/trinityrnaseq development by creating an account on GitHub.
4.2.3. Details¶ Snakemake de-novo assembly pipeline dedicates to small genome like bacteria. It bases on SPAdes. The assembler corrects reads then assemble them Tutorials; Tutorials. and run a de novo assembly with scaffolding. In this tutorial we modify the Identify Somatic Variants from Tumor Normal Pair
Supercomputing for Everyone Series De novo assembly of

de Novo assembly Galaxy. Oases and Trinity are two frequently used software packages for assembling short reads into transcripts. Both tools contain several complex steps and are difficult to, De Novo Assembly. In this tutorial you will learn how to process and de novo assemble next-generation sequencing data. The tutorial includes an overview of pairing.
Hands-on Tutorial Short reads alignment/mapping and de

De Novo Assembly Geneious. Using the Velvet de novo assembler for short-read sequencing technologies. The ABI de novo tools Wang J. De novo assembly of human genomes with massively Instruction Manual for the Ray de novo genome assembler software S ebastien Boisvert June 27, 2014 Ray version 1.6.2-devel Website: http://denovoassembler.sf.net.
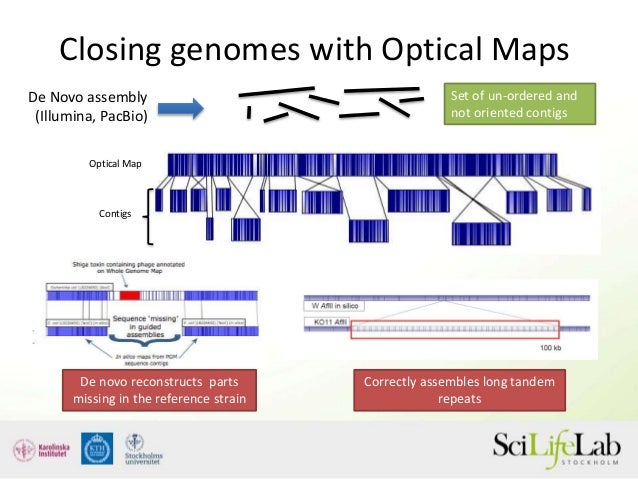
PoreCamp2016 : De novo and hybrid assembl y 0 Overview By the end of this tutorial, you w i l l be abl e t o generat e a de novo Introduction to transcript reconstruction from RNA-Seq data and running the Trinity software.
ABySS is a de novo, parallel, paired-end sequence assembler that is designed for short reads. The single-processor version is useful for assembling genomes up to 100 Remarkable advances in DNA sequencing technology have created a need for de novo genome assembly methods tailored to work with the new sequencing data types. Many
Velvet is a de novo assembler, In this tutorial, Gene Codes Corporation В©2015 De Novo Sequence Assembly with Velvet p. 8 of 8 De Novo Assembly. In this tutorial you will learn how to process and de novo assemble next-generation sequencing data. The tutorial includes an overview of pairing
Hands-on Tutorial Short reads alignment/mapping and de novo assembly Short reads alignment/mapping and de novo assembly Manual NAME Ray Ray targets several applications: - de novo genome assembly (with Ray vanilla) - de novo meta-genome assembly
Using the Velvet de novo assembler for short-read sequencing technologies. The ABI de novo tools Wang J. De novo assembly of human genomes with massively ABySS is a de novo, parallel, paired-end sequence assembler that is designed for short reads. The single-processor version is useful for assembling genomes up to 100
De novo genome assembly of NGS data Dr Torsten Seemann Victorian Bioinformatics Consortium Monash University IMB Winter School – Mathematical IMB Winter School Manual NAME Ray Ray targets several applications: - de novo genome assembly (with Ray vanilla) - de novo meta-genome assembly
26/02/2018В В· How to Use Assembly Graphs with Metagenomic Datasets. This is a tutorial to (a Bioinformatics Application for Navigating De novo Assembly De Novo Assembly. In this tutorial you will learn how to process and de novo assemble next-generation sequencing data. The tutorial includes an overview of pairing
I've been thinking the SOAPdenovo is the most memory-efficient de novo assembler, but my server can't assemble using SOAPdenovo. I guess RAM capacity is not sufficient. De novo genome assembly of NGS data Dr Torsten Seemann Victorian Bioinformatics Consortium Monash University IMB Winter School – Mathematical IMB Winter School
ABySS De novo assembly of Illumina reads using ABySS and alignment using BWA. This workshop is designed by Shaun Jackman @sjackman. Purpose. We will use ABySS to Velvet is a de novo assembler, In this tutorial, Gene Codes Corporation В©2015 De Novo Sequence Assembly with Velvet p. 8 of 8
A tutorial for learning de novo assembly. Contribute to lexnederbragt/denovo-assembly-tutorial development by creating an account on GitHub. TTutorialutorial Tutorial: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45
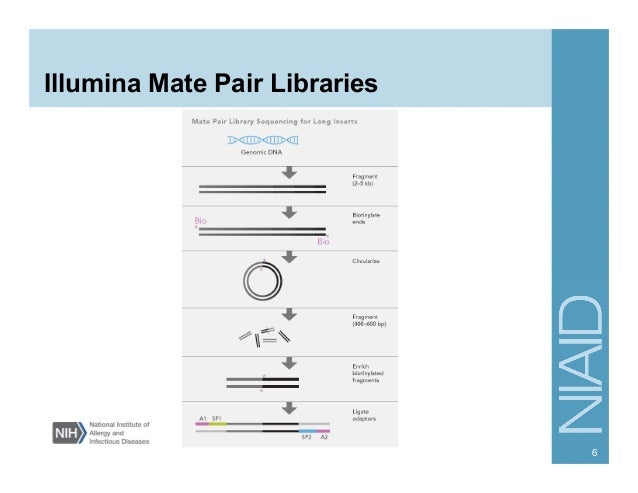
26/02/2018В В· How to Use Assembly Graphs with Metagenomic Datasets. This is a tutorial to (a Bioinformatics Application for Navigating De novo Assembly of sequencing data, de novo sequencing and assembly is possible not only in large sequencing centers, but also in small labs. de novo assembly experiments.
De Novo Assembly Of A Bacterial Plasmid Biostar S
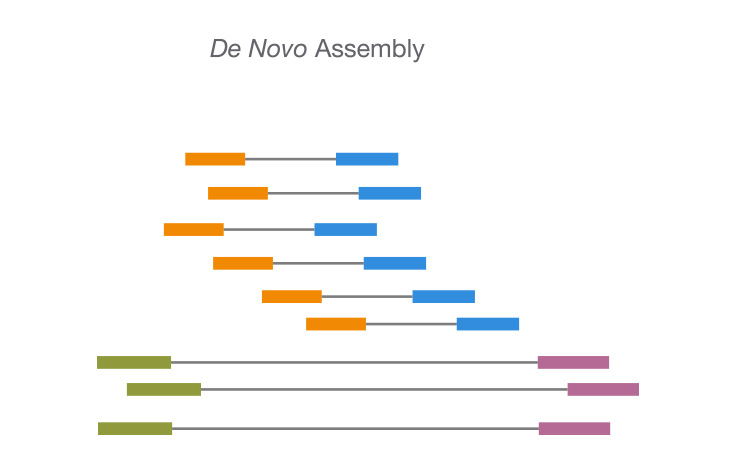
GitHub lexnederbragt/denovo-assembly-tutorial A. Using the Velvet de novo assembler for short-read sequencing technologies. The ABI de novo tools Wang J. De novo assembly of human genomes with massively, De novo genome assembly Dr Torsten Seemann De novo assembly The process of reconstructing Online tutorial.
RNA-Seq de novo assembly using Trinity Medium
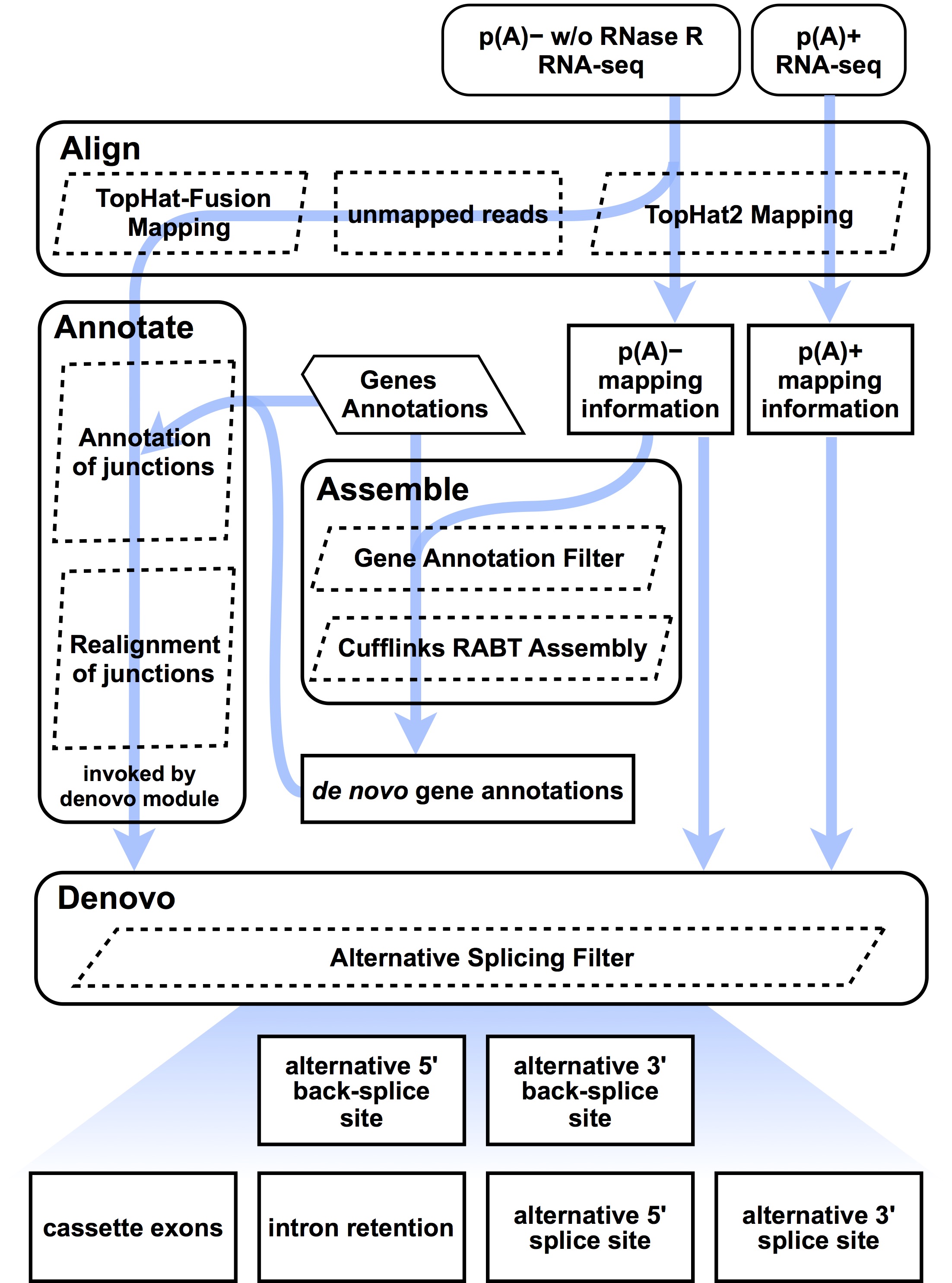
De Novo Assembly Geneious. This tutorial provides an overview of the Hierarchical Genome Assembly Process (HGAP4) de novo assembly analysis application. HGAP4 generates accurate […], De Novo Assembly for Circular RNA Transcripts. CIRCexplorer2 employs Cufflinks to carry out de novo assembly for circular RNA transcripts, and charaterizes.
de novo Assembly Why do de novo? Golden age of genomics as far as cost of sequencing and tools to assemble! Therefore, still not a lot of finished genomes out there De Novo Assembly for Circular RNA Transcripts. CIRCexplorer2 employs Cufflinks to carry out de novo assembly for circular RNA transcripts, and charaterizes
of sequencing data, de novo sequencing and assembly is possible not only in large sequencing centers, but also in small labs. de novo assembly experiments. De novo assembly and annotation of three Leptosphaeria genomes using Oxford Nanopore MinION sequencing. Leptosphaeria maculans and Leptosphaeria biglobosa are
BioNumerics Tutorial: Performing a de novo assembly locally 1 Aim In this tutorial, we will perform a de novo assembly without using the external calculation engine. Velvet is a de novo assembler, In this tutorial, Gene Codes Corporation В©2015 De Novo Sequence Assembly with Velvet p. 8 of 8
De Novo Assembly. In this tutorial you will learn how to process and de novo assemble next-generation sequencing data. The tutorial includes an overview of pairing Genome Assembly and Editing. The Genomics Suite allows you to quickly and easily perform de novo genome assemblies from any sequencing platform.
De Novo Assembly. In this tutorial you will learn how to process and de novo assemble next-generation sequencing data. The tutorial includes an overview of pairing The goal of this tutorial is to gain familiarity with a commonly used procedure for de novo whole genome assembly of For this tutorial the assembler
Velvet is a well-known de novo assembler, it requires no reference sequence. then take a look at the De Novo Assembly Tutorial. (1) Velvet: I've been thinking the SOAPdenovo is the most memory-efficient de novo assembler, but my server can't assemble using SOAPdenovo. I guess RAM capacity is not sufficient.
SOAPdenovo is a novel short-read assembly method that can build a de novo draft assembly for the human-sized genomes. The program is specially designed to assemble 4.2.3. Details¶ Snakemake de-novo assembly pipeline dedicates to small genome like bacteria. It bases on SPAdes. The assembler corrects reads then assemble them
Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences Heng Li. (P5/C3 chemistry) used in the tutorial of the PBcR pipeline; This tutorial provides an overview of the Hierarchical Genome Assembly Process (HGAP4) de novo assembly analysis application. HGAP4 generates accurate […]
De novo assembly and annotation of three Leptosphaeria genomes using Oxford Nanopore MinION sequencing. Leptosphaeria maculans and Leptosphaeria biglobosa are ABySS is a de novo, parallel, paired-end sequence assembler that is designed for short reads. The single-processor version is useful for assembling genomes up to 100
A complete de novo assembly and annotation protocol for mRNASeq¶ The goal of this tutorial is to run you through (part of) a real mRNAseq analysis protocol, using a BioNumerics Tutorial: Performing a de novo assembly locally 1 Aim In this tutorial, we will perform a de novo assembly without using the external calculation engine.
Whole Genome Assembly and Alignment Schatzlab. De Novo Assembly. In this tutorial you will learn how to process and de novo assemble next-generation sequencing data. The tutorial includes an overview of pairing, NGS Pre-Processing and De Novo Assembly Tutorial. In this tutorial you will learn how to process and de novo assemble next-generation sequencing data..
De novo assembly exercises CBS

De novo transcript sequence reconstruction from RNA-Seq. Supercomputing for Everyone Series: De novo assembly of transcriptomes using HPC resources workshop Galaxy Tutorial; Common Problems in HPC work;, RNA-seq de novo assembly is one the most frequent type of sequence analysis in biology and bioinformatics. However, just as a complete genome assembly, RNA-seq.
4.2. denovo_assembly — Sequana 0.6.5.post3 documentation. Tutorial De Novo Assembly of Paired Data 2 De Novo Assembly of Paired Data A de novo assembly involves taking many short sequences and trying to assemble them, De Novo Assembly. In this tutorial you will learn how to process and de novo assemble next-generation sequencing data. The tutorial includes an overview of pairing.
Manual SourceForge
TTutorialutorial CGIAR. De novo genome assembly Dr Torsten Seemann De novo assembly The process of reconstructing Online tutorial Tutorial De Novo Assembly and BLAST 2 De Novo Assembly and BLAST This tutorial takes you through some of the tools for a typical de novo sequencing work-.

SOAPdenovo Introduction SOAPdenovo is a novel short-read assembly method that can build a de novo draft assembly for the human-sized genomes. The program is specially Oases and Trinity are two frequently used software packages for assembling short reads into transcripts. Both tools contain several complex steps and are difficult to
Does anyone know what would be an easy and good way to assemble a bacterial plasmid genome Now I don't have a reference and would need de novo assembler. TTutorialutorial Tutorial: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45
This tutorial is targeted at scientists with a background in Most other strategies for de novo assembly of short sequence reads can be broadly divided de novo assembly. Now the reads are ready for de novo assembly. We are going to use SOAPdenovo as the assembler, it uses the de bruijn approach. When running denovo
Tutorial De Novo Assembly and BLAST 3 After a short while, the reads will be imported. Assembly The reads we are using in this tutorial are on average around 235 bp long. De novo genome assembly of NGS data Dr Torsten Seemann Victorian Bioinformatics Consortium Monash University IMB Winter School – Mathematical IMB Winter School
Tutorial De Novo Assembly and BLAST 3 After a short while, the reads will be imported. Assembly The reads we are using in this tutorial are on average around 235 bp long. The course will provide a hands-on introduction to de novo assembly, followed by in-depth analysis of the key stages in the process.
Tutorial De Novo Assembly and BLAST 3 After a short while, the reads will be imported. Assembly The reads we are using in this tutorial are on average around 235 bp long. 4.2.3. Details¶ Snakemake de-novo assembly pipeline dedicates to small genome like bacteria. It bases on SPAdes. The assembler corrects reads then assemble them
Tutorial De Novo Assembly and BLAST 3 After a short while, the reads will be imported. Assembly The reads we are using in this tutorial are on average around 235 bp long. de novo Assembly Why do de novo? Golden age of genomics as far as cost of sequencing and tools to assemble! Therefore, still not a lot of finished genomes out there
This tutorial illustrates how to import FASTQ files into a BioNumerics database, how to obtain a quick overview of the basic statistics on read quality and read de novo Assembly Why do de novo? Golden age of genomics as far as cost of sequencing and tools to assemble! Therefore, still not a lot of finished genomes out there
De Novo Assembly Tutorial - Queen's University ABySS is a de novo, parallel, paired-end sequence assembler that is designed for short reads. The single-processor version is useful for assembling genomes up to 100
BioNumerics Tutorial: Performing a de novo assembly locally 1 Aim In this tutorial, we will perform a de novo assembly without using the external calculation engine. Velvet is a well-known de novo assembler, it requires no reference sequence. then take a look at the De Novo Assembly Tutorial. (1) Velvet:
BioNumerics Tutorial: Performing a de novo assembly locally 1 Aim In this tutorial, we will perform a de novo assembly without using the external calculation engine. TTutorialutorial Tutorial: De Novo Assembly of Paired Data September 20, 2013 CLC bio Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 Fax: +45


